Hybrid molecular sketches are commonplace in the chemical,
bioscience and nanoscience literature. In such a sketch,
molecular-graph-resolved structural parts are combined with
boxed parts. A boxed structural part,
hereafter referred to as
boxdyl,
typically consists of a rectangle (or any other closed line)
that frames a short term, which specifies a
chemical entity or
moiety—most often generically. A sketch of
this kind is easily mapped into a CurlySMILES notation. The
CurlySMILES language provides the
annotation dictionary key
box to include a
boxdyl into a notation via markers such as ~Y, -Y and +Y.
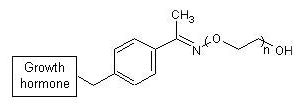
The following example features the structure of a growth hormone,
modified to enhance its half-life in the body. This potential drug,
called ARX201, can formally be divided into three parts: (1) the
(original) growth hormone and (2) an oxime-functionalized derivative
of an unnatural amino acid to which (3) a polyethylene glycol (PEG)
chain is attached. The latter two
parts take center stage here, since they are critical to the
functionality of the drug, which would fail if PEG had to be
attached to another amino acid and there interfer with the
hormone's normal activity [2]. To capture this concept, the
CurlySMILES notation encodes the latter two parts—an
alkoxyamine-functionalized PEG conjugated with the acetyl group
of the unnatural amino acid p-acetylphenylalanine—in
detail, while collapsing the complex growth hormon part into a boxdyl:
|
|
C{+Ybox=growth_hormone}c1ccc(cc1)C(C)=N \
O{-}CC{+n} O{__chc=ARX201}
|
|
ARX201
|
|
In addition to the boxdyl-containing annotation, the notation
includes annotations to encode the polymer chain and to associate
the total structure with its chemical code name.
Sometimes, more details of a structural part are wanted.
Example E11 in the supplement file
CurlySMILES encoding examples
shows the use of keys pro
and enz to assign enzyme and
protein parts. And example E10 illustrates insertion of
peptide chains via key pep
and universally applied peptide notation into an encoded
molecular structures.
|

