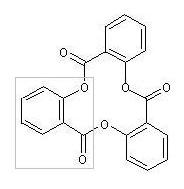
| The CurlySMILES language provides a special encoding format for rings that are based on a structural repeat unit (SRU) such as the trisalicylide, which is built from three salicyl units: | ||||
|
||||
| The salicyl unit is a bivalent group, which can be encoded by employing GEAM annotations: O=C{-}c1ccccc1O{-} . The ring encoding is derived by replacing the second GEAM, which in this example represents the open bond at the phenolic O-atom, by an operational annotation, beginning with the operational annotation marker (OPAM) +r and followed by the entry n=3 specifying the number of repetitions. The OPAM-based format has the advantage that it preserves the principle of the ring design, whose automatic recognition would require elaborate algorithms by a machine interpreter—if exhaustively encoded. Further, this format allows compact encoding of macrocyles with either large SRU numbers or big (or complex) SRU structures. | ||||
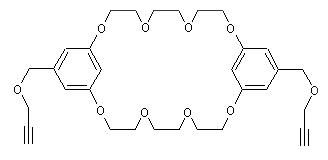
| The following example compares the OPAM-annoted notation with the plain SMILES encoding of a dialkynated bis(m-phenylene)-26-crown-8 (a precursor in the synthesis of cryptands [10.1002/ejoc.200901294]): | ||||
|
||||
Tweet
_
__
__
 __
__
__ ___ 
|
Reference
| [1] | A. Drefahl: CurlySMILES: a chemical language to customize and annotate encodings of molecular and nanodevice structures. J. Cheminf. 2011, 3:1; doi: 10.1186/1758-2946-3-1. |

Custom Search
|